Biomolecular Simulations
Biomolecular simulations make use of computer programs to recreate and visualize the behavior and phenomena that rule biological processes at the molecular level. These tools make it possible to simulate experiments in more controlled conditions than in cells or living organisms, and to perform “theoretical experiments” that would be technically impossible.
These possibilities have resulted in important advances in biomedicine, facilitating the understanding of mechanisms of diseases and the development of drugs, for example. In the Biomolecular Simulations Laboratory, we apply different molecular modelling techniques and simulations to several problems of biomedical interest, such as the stability of viral particles of Zika and Dengue or interactions between proteins that participate in the contraction of the cardiac muscle. These activities are carried out in collaboration with experimental groups in Uruguay and abroad.
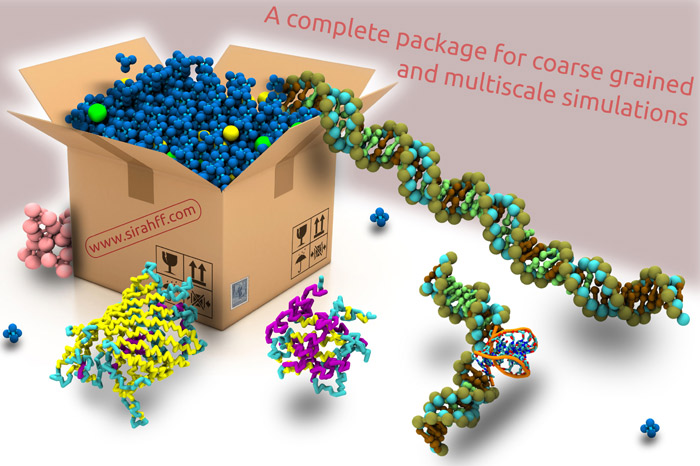
Finally, an important part of our work is dedicated to the development of coarse-grained methods to perform advanced simulations at low computational cost. These methods offer the possibility of improving the comparability of theoretical studies with biochemical / biophysical or molecular biology experiments.
Members




Research lines
Development of the SIRAH coarse-grained force field (Southamerican Initiative for a Rapid and Accurate Hamiltonian).
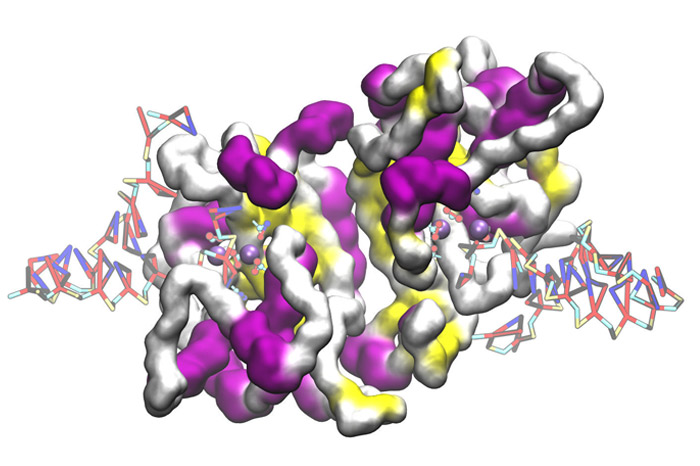
Our group develops and maintains one of the broadest coarse-grained force fields for existing biomolecular simulations. SIRAH (www.sirahff.com) uses a Top Down approach and a classic Hamiltonian, common to atomic force fields. SIRAH is freely distributed with easy-to-use analysis tools, parameters and topologies to simulate DNA, proteins, explicit solvent and phospholipids. Currently, representations are being developed for metal ions, glycans and RNA. This line is entirely developed by our group.
Development of FRET sensors for cyclic nucleotide and redox signalling pathways.
Using bioinformatics and structural modelling, together with coarse-grained simulations, we have developed a new generation of FRET sensors for signaling cAMP, cGMP and redox conditions. These biosensors allow us to reach an unprecedented spatial resolution since they can be genetically fused to the C-terminal of virtually any protein, directing them to any cell compartment. This research line continues with the design of new generations of biosensors in the framework of the ProTeMCA program and in collaboration with foreign experimental groups.
Flavivirus stability studies.
Using multiscale simulations, we study the different factors that affect the stability of viral particles (Virus-like Particles). The reduced computational cost of our simulation scheme allows us to perform comparative simulations of different flaviviruses varying the temperature and pH conditions. The availability of experimental structures of viral particles of Zika, Dengue, Japanese encephalitis (JEV) and tick-borne Encephalitis virus (TBEV) allows us to study the accessibility of different epitopes, helping to understand the mechanisms of viral neutralization by antibodies. Additionally, the computational approach enables the identification of amino acids involved in the acid firing mechanism of flaviviruses, which could contribute significantly to the development of vaccines through the creation of attenuated viruses. These studies are part of collaborations with national and foreign experimental groups.
Courses and congresses
- “Latin American Initiative for Molecular Simulations” Expert´s meeting. Nov. 2018. IP Montevideo.
- OpenLab “Performing Molecular Simulations with the Sirah force field”. 2015 and 2017 editions, IP Montevideo. Organizer: Sergio Pantano.
- VIII PostLATAM course Membrane Lipids, Transporters, Channels…and all that crosstalk, Nov. 2015, Salto, Uruguay.
- Joint meeing SAB/SBFUy “Latin American Crosstalk in Biophysics and Physiology”, Nov. 2015, Salto, Uruguay.
- “Introduction to Structural Biology and Bioinformatics”, Nov. 2013. IP Montevideo.
- Course and workshop “Ion Channels: From Molecules to Pathology”, April 2012. Universidad de la República – IP Montevideo.
- Course “NFS Workshop on Multiscale Modeling and Simulation”, Set. 2012, IP Montevideo.
- “Hands-on Course: Coarse Grain Methods for Biomolecular Simulations”, Set 2011, IP Montevideo.
- Course and workshop “Computational Modelling and Simulation of Biological Systems”, February 2010, IP Montevideo.
- “Conference on Molecular Aspects of Cell Biology: A Perspective from Computational Physics”, Oct. 2010. International Centre for Theoretical Physics (ICTP), Trieste, Italy.
Projects
2018-2019 – “Modulation of G-protein coupled receptors through molecular dynamics simulations as a tool for the rational planning of new drugs”. Responsible: Hugo Verli-Sergio Pantano. LNCC, Santos Dumont, Brasil.
2017-2018 – “In silico characterization of drug targets for Zika and Dengue”. Responsible: Gustavo Seabra-Sergio Pantano. LNCC, Santos Dumont, Brasil.
2018-2020 – “Molecular mechanism of signaling in bacteria: directionality from signal to response”. Responsible: Alejandro Buschiazzo. Fondo Clemente Estable, ANII.
2015-2018 – “Design of biosensors for simultaneous monitoring of redox signaling and cAMP: From the computer to the cell and back to the computer”. Responsible: Sergio Pantano. Fondo María Viñas, ANII.
Main publications
vacio
2023
- Ulinici M, Soñora M, Orsini E, Licastro D, Dal Monego S, Todiras M, Lungu L, Groppa S, Marcello A. Genome Sequences of SARS-CoV-2 Strains from the Republic of Moldova. Microbicrobiol Resour Announc. 2023 Jan 24;12(1):e0113222. doi: 10.1128/mra.01132-22.
2022
- Sanguinetti M, Silva Santos LH, Dourron J, Alamón C, Idiarte J, Amillis S, Pantano S, Ramón A. Substrate Recognition Properties from an Intermediate Structural State of the UreA Transporter. Int. J. Mol. Sci. 2022, 23,16039.
- Rueda AJV, Conteville L, Pantano S, González W, Women in bioinformatics & data science – Latin America, MethodsX, 2022, 9:101907 eCollection 2022.
- Pantano S, Back and forth modeling through biological scales. BBRC, 2022, 633, 39.
- Acevedo M, Alonso-Palomares L, Montes de Oca M, Bustamante A, Gaggero A, Paredes F, Cortés C, Pantano S, Navarrete M, Valiente-Echeverría F, Soto-Rifo R, Neutralization of the emerging SARS-CoV-2 variant Lambda by antibodies elicited by inactivated virus and mRNA vaccines, Nature Microbiol. 2022, 7 (4), 524. – Author correction at doi: 10.1038/s41564-022-01154-4.
- Soñora M, Barrera EE, Pantano S, The stressed life of a lipid in the Zika Virus membrane, BBA – Membranes, 2022, 1864 (1), 183804.
- Pantano S, Share data, share methods, share science, MethodsX, 2022, 9: 101607.
- Santana E, de Paula B, Valvassori L, Pereira CD, Santos LH, Campos S, Franco O, Morais L, Torquato M, Synoeca-MP: New insights into its mechanism of action by using NMR and molecular dynamics simulations approach. Peptide Science, 2022, DOI:10.1002/pep2.24293
- Santos LH, Kronenberger T, Almeida RG, Silva EB, Rocha REO, Oliveira JC, Barreto LV, Skinner D, Fajtová P, Giardini MA, Woodworth B, Bardine C, Lourenço AL, Craik CS, Poso A, Podust LM, McKerrow JH, Siqueira-Neto JL, O’Donoghue AJ, da Silva Júnior EN, Ferreira RS. Structure-Based J Chem Inf Model. 2022,62,6553-6573. doi: 10.1021/acs.jcim.2c00693.
2021
- Buratto D, Saxena A, Ji Q, Yang Q, Pantano S, Zonta F,Rapid assessment of binding affinity of SARS-COV-2 spike protein to the human angiotensin-converting enzyme 2 receptor and to neutralizing biomolecules based on computer simulations, Frontiers Immunol. 2021, 12: 730099.
- BesadaP, Gallardo-Gómez M, Pérez-Márquez T, Patiño-Álvarez L, Pantano S, Silva-López C, Terán C, Arévalo-Gómez A, Ruz-Zafra A, Fernández-Martín J, Ortolano S, The new pharmacological chaperones PBXs increase α-Galactosidase A activity in Fabry disease cellular models, Biomolecules, 2021, 11, 1856.
- González-Puelma J, Aldridge J, Montes de Oca M, Pinto M, Uribe-Paredes R, Fernández-Goycoolea J, Alvarez-Saravia D, Álvarez H, Encina G, Weitzel T, Muñoz R, Olivera-Nappa Á, Pantano S, Navarrete MA. Mutation in a SARS-CoV-2 Haplotype from Sub-Antarctic Chile Reveals New Insights into the Spike’s Dynamics. Viruses, 2021, 13(5):883
- Barrera EE, Pantano S, Zonta F, A homogeneous dataset of polyglutamine and glutamine rich aggregating peptides simulations, DiB, 2021, 36:107109.
- Machado MR, Pantano S, Fighting viruses with computers, right now, Curr. Op. Virology, 2021, 48:91.
- Barrera EE, Zonta F, Pantano S, Dissecting the role of glutamine in seeding peptide aggregation. CSBJ, 2021, 19: 1595.
- Asciutto EK, Pantano S, General IJ, Physical Interactions Driving the Activation/Inhibition of Calcium/Calmodulin Dependent Protein Kinase II, Jmol. Graph. and Modelling, 2021, 107875
- Klein F, Sardi F, Machado MR, Ortega CK, Comini M, Pantano S, CUTie2: The Attack of the cyclic nucleotide sensor clones. Frontiers in Molecular Biosciences, 2021,18: 48
- Garay P, Barrera EE, Klein F, Machado MR, Soñora M, Pantano S, The SIRAH-CoV-2 Initiative: A coarse-grained simulations’ dataset of the SARS-CoV-2 proteome. Frontiers in Medical Technology, 2021, 3:644039.
- Soñora M, Martínez L, Pantano S, Machado MR, Wrapping Up Viruses at Multiscale Resolution: Optimizing PACKMOL and SIRAH Execution for Simulating the Zika Virus. JCIM, 2021, 61:408
- Klein F, Barrera EE, Pantano S, Assessing SIRAH’s Capability to Simulate Intrinsically Disordered Proteins and Peptides, 2021, JCTC, 61:408
- Aldunate F, Echeverría N, Chiodi D, López P, Sánchez-Cicerón A, Soñora M, Cristina J, Moratorio G, Hernández N, Moreno P. Resistance-associated substitutions and response to treatment in a chronic hepatitis C virus infected-patient: an unusual virological response case report. BMC Infect Dis. 2021 Apr 26;21(1):387. doi: 10.1186/s12879-021-06080-0.
2020
- Marchetto A, Chaib ZS, Rossi CA, Ribeiro R, Pantano S, Rossetti G, Giorgetti A, CGMD Platform: Integrated Web Servers for the Preparation, Running, and Analysis of Coarse-Grained Molecular Dynamics Simulations. Molecules. 2020, 25:5934.
- Ahyayauch H, García-Arribas AB, Masserini ME, Pantano S, Goñi FM, Alonso A, β-amyloid (1-42) peptide adsorbs but does not insert into ganglioside-containing phospholipid membranes in the liquid-disordered, IJBM, 2020,164:2651.
- Medeiros A, Benítez D, Korn RS, Ferreira VC, Barrera E, Carrión F, Pritsch O, Pantano S, Kunick C, de Oliveira CI, Orban OCF, Comini MA. Mechanistic and biological characterisation of novel N5-substituted paullones targeting the biosynthesis of trypanothione in Leishmania. J Enzyme Inhib Med Chem. 2020, 35:1345.
- Klein F, Caceres-Rojas D, Carrasco M, Tapia JC, Caballero J, Alzate-Morales JH, Pantano S, Coarse-Grained parameters for divalent cations within the SIRAH force field. JCIM, 2020, JCIM. 2020, 60:3935.
- Garduno-Robles A, Alata M, Piazza V, Cortes Sanchez C, Eguibar JR, Pantano S, Hernandez VH, MRI features in a rat model of H-ABC tubulinopathy. Frontiers in Neuroscience, 2020, 14:555.
- Frigini EN, Barrera EE, Pantano S, Porasso RD. Role of membrane curvature on the activation/deactivation of Carnitine Palmitoyltransferase 1A: A coarse grain molecular dynamic study. BBA Biomembranes. 2020, 1862, 2, 183094.
- Herrera MG, Gómez Castro MF, Prieto E, Barrera E, Dodero VI, Pantano S, Chirdo F. Structural conformation and self‐assembly process of p31‐43 gliadin peptide in aqueous solution. Implications for celiac disease. FEBS 2020, 287:2134.
- Machado M, Pantano S, Split the Charge Difference in Two! a Rule of Thumb for Adding Proper Amounts of Ions in MD Simulations. JCTC, 2020, 16:1367.
- Garay P, Barrera E, Pantano S, Post-translational modifications at the coarse-grained level with the SIRAH force field. Journal of Chemical Information and Modeling, 2020, JCIM, 2020, 60:964.
2019
- Pantano S. Editorial Comment on Dissecting the role of glycosylation in Zika virus envelope with biochemical methods. Biochem Biophys Res Commun. 2019, 520:690
- Otero L, Martínez-Rosales C, Barrera E, Pantano S, Salinas G.Complex I and II Subunit Gene Duplications Provide Increased Fitness to Worms. Front Genet. 2019, 10:1043.
- Barrera E, Chirdo F, Pantano S. Commentary: p31-43 Gliadin Peptide Forms Oligomers…Frontiers in Immunology, 2019, DOI: 10.3389/fimmu.2019.02792
- Chao YC, Surdo NC, Pantano S, Zaccolo M. Imaging cAMP nanodomains in the heart. Biochem Soc Trans. 2019 Oct 31. pii: BST20190245. doi: 10.1042/BST20190245.
- Herrera MG, Gómez Castro MF, Prieto E, Barrera E, Dodero VI, Pantano S, Chirdo F. Structural conformation and self-assembly process of p31-43 gliadin peptide in aqueous solution. Implications for celiac disease FEBS J. 2019 Oct 28. doi: 10.1111/febs.15109
- Barrera E, Machado M, Pantano S, Fat SIRAH: Coarse-Grained Phospholipids To Explore Membrane−Protein Dynamics. JCTC, 2019, 15:10.
- Machado M, Barrera E, Klein F, Sonora M, Silva S, Pantano S, The SIRAH force field 2.0: Altius, Fortius, Citius.JCTC, 2019, 15:2719.
- Piattoni CV, Sardi F, Klein F, Pantano S, Bollati-Fogolin M, Comini M. New red-shifted fluorescent biosensor for monitoring intracellular redox changes. Free Radical Biology and Medicine, 2019, 134:545.
- Riccardi E. Pantano S, Potestio R. Envisioning data sharing for the biocomputing community. Interface Focus, 2019, s. 9(3),20180005.
- Machado M, Zeida A, Darré L, Pantano S. From quantum to subcellular scales: multiscale simulation approaches and the SIRAH force field. 2019, Interface Focus, 2019, s. 9(3),20180085.
- Gómez Castro, MF, Miculán E, Herrera MG, Ruera C, Perez F, Prieto ED, Barrera E, Pantano S,, Carasi P, Chirdo FG. p31-43 Gliadin Peptide Forms Oligomers and Induces NLRP3 Inflammasome/Caspase 1- Dependent Mucosal Damage in Small Intestine. Frontiers in Immunology, 2019, 10:31.
2018
- Tosar JP, Gambaro F, Darré L, Westhof E, Cayota A, Dimerization confers increased stability to nucleases inextracellular 5’ halves from glycine and glutamic acid tRNAs. NAR, In press. DOI: 10.1093/nar/gky495
- Zonta F, Buratto D, Crispino G, Carrer A, Bruno F, Yang G, Mammano F and Pantano S. Cues to opening mechanisms from in silico electric field excitation of Cx26 hemichannel and in vitro mutagenesis studies in HeLa transfectans. Frontiers in Molecular Neuroscience. 2018, vol 11, art 170.
- Esteban C, Donati I, Pantano S, Villegas M, Benegas J, Paoletti S, Dissecting the conformational determinants of Chitosan and Chitlac oligomers. Biopolymers, 2018, e23221.
- Viso JF, Belelli P, Machado M, González H, Pantano S, Amundarain MJ, et al. (2018) Multiscale modelization in a small virus: Mechanism of proton channeling and its role in triggering capsid disassembly. PLoS Comput Biol 2018, 14(4): e1006082.
- Sulpizi M, Faller R, Pantano S. Multiscale modeling on biological systems. BBRC. 2018, 498:263.
- Brandner AF, Schueller A, Melo F, Pantano S. Exploring DNA dynamics within oligonucleosomes with coarse-grained simulations: SIRAH force field extension for protein-DNA complexes. BBRC, 2018, 498:319
2017
- Machado M, Gonzalez HC, Pantano S. MD Simulations of Virus-Like Particles with Supra CG solvation affordable to desktop computers. JCTC. 2017, 13: 5106.
- Marcello A, Pantano S. Interdisciplinary approaches to the study of flavivirus. BBRC, 2017, 492:531.
- Barrera E, Frigini EN, Porasso RD, Pantano S. Modeling DMPC lipid membranes with SIRAH force-field. Journal of Molecular Modelling. 2017, 23: 259
- Surdo N, Berrera M, Koschinski A, Brescia M, Machado M, Carr C, Morotti S, Grandi E, Wright P, Bers D, Gorelik J, Pantano S, Zaccolo M. FRET biosensor uncovers cAMP nano-domains at β-adrenergic targets that dictate precise tuning of cardiac contractility. Nature Comm, 2017, 8: 15031.
- Festari MF,Trajtenberg F, Berois N, PantanoS , Revoredo L, Kong Y, et al. Revisiting the human polypeptide GalNAc-T1 and T13 paralogs. Glycobiology. 2017, 27:140.
2016
- Astrada S, Gomez Y, Obal G, Pritsch O, Vallespí MG, Bollati-Fogolín M. Comparative analysis reveals amino acids critical for anticancer activity of peptide CIGB-552. J. Pep. Sci. 2016, 22:711.
- Calì T, Frizzarin M, Luoni L, Zonta F, Pantano S, Cruz C, Bonza MC, Bertipaglia I, Ruzzene, De Michelis MI, Damiano N, Marina O, Zanni G, Zanotti G, Brini M, Lopreiato R, Carafoli E. The ataxia related G1107D mutation of the plasma membrane Ca2 + ATPase isoform 3 affects its interplay with calmodulin and the autoinhibition process. BBA – Mol. Basis Dis. 2016, 1863:165.
- Machado MR and Pantano S. SIRAH Tools: mapping, backmapping and visualization of coarse-grained models. Bioinformatics, 2016, 32:1568.
2015
- Machado MR and Pantano S. Exploring LacI−DNA Dynamics by Multiscale Simulations Using the SIRAH force field. JCTC, 2015, 11:5012.
- Jäger AV, De Gaudenzi JG, Mild JG, Cormack BM, Pantano S, Altschuler DL, Edreira MM. Identification of novel cyclic nucleotide binding proteins in Trypanosoma cruzi. Mol Biochem Parasitol. 2015, 198:104.
- Darré L, Machado MR, Brandner AF, Ferreira S, Gonzalez HC, Pantano S. SIRAH: a structurally unbiased coarse-grained force field for proteins with aqueous solvation and long-range electrostatics. JCTC, 2015, 11:723.
- Morande PE, Borge M, Abreu C, Galletti J, Zanetti SR, Nannini P, Bezares RF, Pantano S, Dighiero G, Oppezzo P, Gamberale R, Giordano M. Surface localization of high-mobility group nucleosome-binding protein 2 (HMGN2) on leukemic B cells from chronic lymphocytic leukemia patients is related to secondary autoimmune hemolytic anemia. Leuk Lymphoma. 2015. Jan 21:1-8.
2014
- Sanguinetti M, Amillis S, Pantano S, Scazzocchio C and Ramón A. Modeling and mutational analysis of Aspergillus nidulans UreA, a member of the subfamily of urea/H+ transporters in fungi and plants. Open Biology, 2014, 4:140070
- Zecchin A, Pattarini L, Gutierrez MI, Mano M, Mai A, Valente S, Myers MP, Pantano S, and Giacca M. Reversible acetylation regulates vascular endothelial growth factor receptor-2 activity. Journal of Molecular Cell Biology, 2014, 6:116.
2013
- Gonzalez HC, Darré, L. Pantano, S. Transferable Mixing of Atomistic and Coarse-Grain Water Models. J. Phys. Chem. B, 2013, 117 :14438.
- Pantano S, Montecucco C. The Blockade of the Neurotransmitter Release Apparatus by Botulinum Neurotoxins. Cell. Mol. Life Sci. 2013, DOI:10.1007/s00018-013-1380-7.
- Megighian A, Zordan M, Pantano S, Scorzeto M, Rigoni M, Zanini D, Rossetto O, Montecucco C. Evidence for a radial SNARE super-complex mediating neurotransmitter release at the Drosophila neuromuscular junction. J. Cell. Sci., 2013, 136: 3134.
- Almeida RS, Loss O, Colabardini AC, Brown NA, Bignell E, Savoldi M, Pantano S, Goldman MH, Arst HN Jr, Goldman GH. Genetic Bypass of Aspergillus nidulans crzA Function in Calcium Homeostasis. G3 (Bethesda), 2013, 3:1129.
Book chapters
- David A Case, H Metin Aktulga, Kellon Belfon, Ido Ben-Shalom, Scott R Brozell, David S Cerutti, Thomas E Cheatham III, Vinícius Wilian D Cruzeiro, Tom A Darden, Robert E Duke, George Giambasu, Michael K Gilson, Holger Gohlke, Andreas W Goetz, Robert Harris, Saeed Izadi, Sergei A Izmailov, Chi Jin, Koushik Kasavajhala, Mehmet C Kaymak, Edward King, Andriy Kovalenko, Tom Kurtzman, Taisung Lee, Scott LeGrand, Pengfei Li, Charles Lin, Jian Liu, Tyler Luchko, Ray Luo, Matias Machado, Viet Man, Madushanka Manathunga, Kenneth M Merz, Yinglong Miao, Oleg Mikhailovskii, Gérald Monard, Hai Nguyen, Kurt A O’Hearn, Alexey Onufriev, Feng Pan, Sergio Pantano, Ruxi Qi, Ali Rahnamoun, Daniel R Roe, Adrian Roitberg, Celeste Sagui, Stephan Schott-Verdugo, Jana Shen, Carlos L Simmerling, Nikolai R Skrynnikov, Jamie Smith, Jason Swails, Ross C Walker, Junmei Wang, Haixin Wei, Romain M Wolf, Xiongwu Wu, Yi Xue, Darrin M York, Shiji Zhao, Peter A Kollman. Amber 21. University of California, San Francisco, 2021.
- Klein F, Abreu C, Pantano S. How to make the CUTiest sensor in 3 simple steps for computational pedestrians. In cAMP signalling, Methods in Molecular Biology, Zaccolo, M et al. Eds. Springer International Publishing, 2022, 2483; pp 255-264.
- Barrera E, Pantano S. Simulating Transmembrane Proteins with the Coarse-Grained SIRAH Force Field: Tips and Tricks for Setting Up and Running in AMBER. Chapter 3 in A Practical Guide to Recent Advances in Multiscale Modeling and Simulation of Biomolecules. AIP, 2023