Functional Genomics
Our scientific goal is to clarify the biological role of small regulatory RNAs in the realm of human cancer. Moreover, we work in close collaboration with the University Hospital and the National Cancer Program, to which we provide technological and experimental support for Clinical Oncology research, as well as developing new diagnostic biomarkers for cancer and disease.
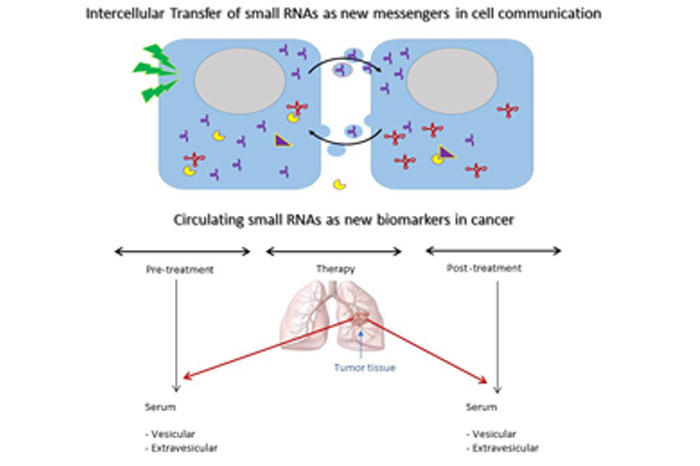
In recent years, our main research goals focused on the study of a new class of molecules called small non-coding RNAs. Our work shows that fragmentation of these RNAs generates molecules capable of regulating cell survival pathways related to cell stress and proliferation. We primarily focus on fragments derived from transfer RNAs and Y-RNAs and their secretion from normal tumor cells. We study how other cells are able to receive and sense these RNAs that were previously released onto the extracellular environment. We evaluate these pathways as a new communication mechanism between cells. This research and other lines of work at Functional Genomics aim at identifying new molecular pathways in cancer initiation and progression with special emphasis on new therapeutic targets and diagnostic biomarkers.
Members










Research lines
Small RNAs derived from tRNA and Y-RNAs as new molecular pathways in cancer and their potential as a source of new diagnostic biomarkers in cancer.
Our main line of research is oriented to the role of new classes of small regulatory RNAs derived from tRNA and Y-RNA as central actors in the proliferation and survival responses to stress, and its potential as new molecular mechanisms in the initiation and progression of cancer. The focus of our current research activities is on the extracellular biology of these regulatory RNAs, and their ability to work as signaling molecules between cells.
Part of our work has shown that these small RNAs are actively secreted by cells through extracellular vesicles or extra-vesicular fractions, being transferred to other cells, constituting a new mechanism of intercellular communication and transfer of genetic information. Additionally, its unique stability in the extracellular environment allows its detection in biological fluids, positioning itself as potential molecular biomarkers in human cancer.
Currently, in collaboration with the Clinical Oncology Service of the Hospital de Clínicas and the National Cancer Institute, we are studying the diagnostic value of small circulating RNAs derived from tRNAGlu, tRNAGly and Y4-RNA in patients with lung cancer.
Research and Development activities with health centers.
Our laboratory is currently participating in a series of initiatives to incorporate new diagnostic biomarkers in oncology, as well as genomic and molecular tools.
Genetic susceptibility to breast cancer. Together with the Hospital de Clínicas of the Faculty of Medicine (Udelar), we have incorporated in the oncology routine the study of mutations of a panel of 11 hereditary predisposition genes to breast and ovarian cancer. This procedure uses next-generation deep sequencing for analysis including the BRCA1 and BRCA2 genes among others.
Biomarkers of therapeutic prediction in lung cancer. Our laboratory performs fluorescent in situ hybridization (FISH) assays to detect translocations of the ALK gene, as a way to predict sensitivity to treatment with inhibitors (Crizotinib) in lung cancer.
Courses
- International course “Deciphering regulator RNA functions by high-throughput sequencing”. December 4-8, 2017. Organizer: Dr. Cayota. Funded by: UNU-BIOLAC, FOCEM and private sponsors.
Projects
2016-2018 – “Implementation of genetic tests for breast cancer risk by deep sequencing of BRCA1 and BRCA2 genes in Uruguayan women”. Fondo María Viñas – ANII.
2017-2019 – “Biosensors for the decentralized detection of exosomes and Dengue virus”. Responsible: Juan Pablo Tosar. Funded by: CSIC, University of the Republic.
2017-2019 – “tRNA-derived small RNAs as mediators of survival and growth signals”. Responsible: Alfonso Cayota. Funded by: CSIC, University of the Republic.
2009-2010 – Member of the Uruguayan team in the Multicenter Pilot Project on Breast Cancer in Latin America with the National Cancer Institute of the United States and its counterparts in Brazil, Argentina, Mexico, Chile and Uruguay. Participants: National Cancer Control Program – Ministry of Public Health; Faculty of Medicine (Udelar), Cancer Research Program of the Institut Pasteur de Montevideo.
2009-2010 – “Helicasas with chromodomain in the initiation and progression of leukemic processes”. Fondo Clemente Estable – ANII.
2009-2010 – “MicroRNA-dependent chromohelicases as a novel tumorigenic pathway in human cancer”. The Pasteur – Weizmann Joint Research Program.
2008-2010 – “Identification of new molecular mechanisms of human carcinogenesis mediated by micro-RNAs and chromatin remodeling proteins”. Sectorial Commission for Scientific Research (CSIC).
2008-2009 – “miR-181 gene analysis as molecular marker in the initiation and progression of Chronic Lymphoid Leukemia”. Honorary Committee Against Cancer (CHLCC), Uruguay.
Main publications
vacio
2018
- Fromm, B; Kang, W; Rovira, C; Cayota, A; Witwer, KW; Friedländer, M and Tosar, J.P. (2018): Plant microRNAs in human sera are likely contaminants. Journal of Nutritional Biochemistry (In press).
- Fromm, B.; Tosar, J.P.; Yu, L.; Halushka, M.; Witwer, K. (2018) miR-21-5p and miR-30a-5p are identical in human and bovine, have similar isomiR distribution, and cannot be used to identify xenomiR uptake from cow milk. Journal of Nutrition. Accepted manuscript, (In press).
- Tosar, J.P., Gambaro, F., Darre, L., Pantano, S., Westhof, E. and Cayota, A. (2018) Dimerization confers increased stability to nucleases in 5′ halves from glycine and glutamic acid tRNAs. Nucleic Acids Res. gky495; doi: 10.1093/nar/gky495
- Tosar, J.P., Rovira, C. and Cayota, A. (2018) Non-coding RNA fragments account for the majority of annotated piRNAs expressed in somatic non-gonadal tissues. Communications Biology, 1, 2.
- Tosar, J.P. and Cayota, A. (2018) Detection and Analysis of Non-vesicular Extracellular RNA. Methods Mol Biol, 1740, 125-137.
- Fagúndez, P.; Brañas, G.; Cairoli, E.; Laíz, J. andTosar, J.P. (2018) An electrochemical biosensor for rapid detection of anti-dsDNA antibodies in absolute scale. Analyst, 143, 3874-3882
2017
- Tosar, J.P., Cayota, A., Eitan, E., Halushka, M.K. and Witwer, K.W. (2017) Ribonucleic artefacts: are some extracellular RNA discoveries driven by cell culture medium components? J Extracell Vesicles, 6, 1272832.
- Mateescu, B., Kowal, E.J., van Balkom, B.W., Bartel, S., Bhattacharyya, S.N., Buzas, E.I., Buck, A.H., de Candia, P., Chow, F.W., Das, S. et al. (2017) Obstacles and opportunities in the functional analysis of extracellular vesicle RNA – an ISEV position paper. J Extracell Vesicles, 6, 1286095.
2016
- Doldán, X., Fagúndez, P., Cayots, A., Laíz, J., Tosar, J.P. (2016) Electrochemical sandwich immunosensor for determination of exosomes based on surface marker-mediated signal amplification. Anal Chem, 88, 10466-10473
2015
- Tosar, J.P., Gambaro, F., Sanguinetti, J., Bonilla, B., Witwer, K.W. and Cayota, A. (2015) Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res, 43, 5601-5616.
- Cairoli, E., Danese, N., Teliz, M., Bruzzone, M.J., Ferreira, J., Rebella, M. and Cayota, A. (2015) Cumulative dose of hydroxychloroquine is associated with a decrease of resting heart rate in patients with systemic lupus erythematosus: a pilot study. Lupus, 24, 1204-1209.
2014
- Tosar, J.P., Rovira, C., Naya, H. and Cayota, A. (2014) Mining of public sequencing databases supports a non-dietary origin for putative foreign miRNAs: underestimated effects of contamination in NGS. RNA, 20, 754-757.
- Garcia-Silva, M.R., Cabrera-Cabrera, F., das Neves, R.F., Souto-Padron, T., de Souza, W. and Cayota, A. (2014) Gene expression changes induced by Trypanosoma cruzi shed microvesicles in mammalian host cells: relevance of tRNA-derived halves. Biomed Res Int, 2014, 305239.
- Garcia-Silva, M.R., das Neves, R.F., Cabrera-Cabrera, F., Sanguinetti, J., Medeiros, L.C., Robello, C., Naya, H., Fernandez-Calero, T., Souto-Padron, T., de Souza, W. et al. (2014) Extracellular vesicles shed by Trypanosoma cruzi are linked to small RNA pathways, life cycle regulation, and susceptibility to infection of mammalian cells. Parasitol Res, 113, 285-304.
- Garcia-Silva, M.R., Sanguinetti, J., Cabrera-Cabrera, F., Franzen, O. and Cayota, A. (2014) A particular set of small non-coding RNAs is bound to the distinctive Argonaute protein of Trypanosoma cruzi: insights from RNA-interference deficient organisms. Gene, 538, 379-384.