Scientists from the Institut Pasteur in Paris and Montevideo have made a significant breakthrough in understanding how bacteria like those responsible for tuberculosis, leprosy, and diphtheria multiply and grow. Their research, published in Nature Microbiology, sheds light on two new proteins critical to this process. This newfound knowledge could pave the way for developing better tools to combat these pathogens.
This work, leaded by Ann Marie Wehenkel and Rosario Durán from the Structural Microbiology Unit (Institut Pasteur Paris) and the Analytical Biochemistry and Proteomics Unit (Ubypa, Institut Pasteur de Montevideo and Instituto Clemente Estable), respectively, not only enhances our understanding of bacterial growth and division but also opens exciting new directions of research with potential implications for the development of novel antimicrobial drugs.
At a glance, bacteria multiply by growing to a certain size and then dividing into two identical cells. On a molecular level, this growth and division are orchestrated by two complex multi-protein machineries known as the elongasome and divisome. While much is known about these systems in model bacteria, there are still many important bacterial groups for which this information remains a mystery.
One such group is Corynebacteriales, which includes dangerous human pathogens like Mycobacterium tuberculosis (causing tuberculosis), Mycobacterium leprae (causing leprosy), and Corynebacterium diphtheriae (causing diphtheria). It also features industrially significant bacteria like Corynebacterium glutamicum, used to produce glutamic acid for flavoring and food additives.
However, little is known about the growth and division mechanisms of these bacteria, mainly due to their unique characteristics, such as polar growth and complex cell walls. .
The research originally aimed to uncover the missing pieces of the puzzle, namely to identify and characterize the proteins responsible for cell division and growth in these bacteria.
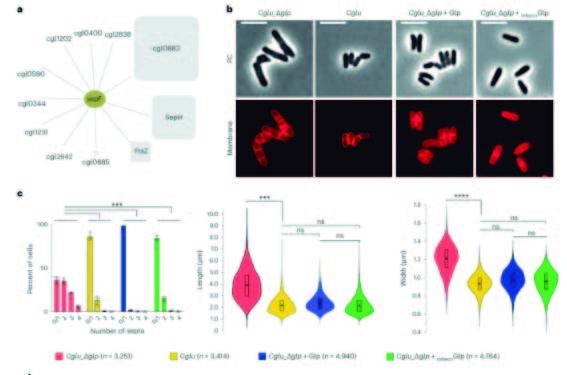
This collaborative effort between the Pasteur Institutes in Paris and Montevideo has revealed for the first time a direct link between the elongation and division machineries. This connection is assured by two new key players in the divisome: Glp, a molybdotransferase, and GlpR, its membrane receptor, that play a crucial role in the bacterial cell cycle. These proteins act as a bridge, facilitating the transition from the late divisome in dividing cells to the polar elongasome in the new daughter cells.
This research also unveils an intriguing feature of this bacterial group: the enzyme Glp, originally responsible for molybdenum processing, has experienced ‘evolutionary repurposing’, a remarkable transformation meaning that Glp’s initial role (molybdotransferase) has evolved over time to take on a new function in an entirely distinct biological process (orchestrating a protein network that govern cell division).
Strikingly, this phenomenon echoes what is found in the nervous system of higher eukaryotes, where a similar enzyme called gephyrin, also a molybdotransferase, has adapted to bind and organize GlyR receptors that play a pivotal role in synaptic signaling. Evolutionary repurposing of enzymes, where they adapt to perform different tasks than their original role, showcases one of life’s incredible abilities to respond to changing conditions. This work not only enhances our understanding of bacterial growth and division but also opens up new exciting directions of research with potential implications for the development of novel antimicrobial drugs.